Microbial Warriors: A New Hope in the Fight Against Cancer
Cancer is a complex disease that requires a multifaceted approach to treatment in order to prevent regression. In the past, traditional treatments in oncology have consisted of small molecule cytotoxic drugs (alkylating agents, antimetabolites, etc.), and recently, more targeted therapies like tyrosine kinase inhibitors (TKIs), monoclonal antibodies (mABs), antibody-drug conjugates (ADCs), bi-specific T cell engagers (BITEs), etc. However, resurgence of a previously underutilized field of cancer therapy has been making a comeback: microbial-based therapies, aka “bugs as drugs.”
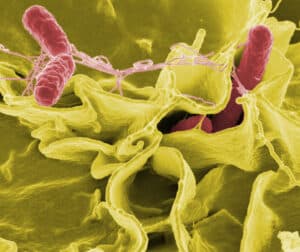
Bacteria have an advantage over molecule-based therapies in that they can be used to target hypoxic tumors and potentially offer better tissue penetration. In addition, they can be genetically modified to enhance targeting, carry or produce different payloads, and to overcome drug resistance. The most common mechanisms of action of microbial-therapy are direct immunotherapeutic (in which the bacteria elicit an anti-tumor immune response), as vectors (to ferry therapeutics or produce them within the tumor environment), and through production of bacterial toxins and enzymes [1-4]. Some of the commonly used bacterial genera include Salmonella (shown in Figure 1 below), Clostridium, Bifidobacterium, Lactobacillus, Escherichia, Pseudomonas, Caulobacter, Listeria, Proteus, and Streptococcus.
(Figure 1. Scanning electron micrograph of Salmonella Typhimurium, a common food-borne pathogen that has also been used as a novel anti-tumor treatment)
The immunotherapeutic effect of bacteria stems from their ability to naturally release pathogen-associated molecular patterns (PAMPs), which leads to the release pro-inflammatory cytokines and a Th1 cytotoxic cell response. Additional genetically engineered effectors, such as the production and subsequent MHC class 1 presentation of highly antigenic proteins (e.g., ovalbumin), can enhance the body’s immune response to the target tumor. Many bacteria used in microbial therapeutics—such as Salmonella, Escherichia, and Clostridium—are also capable of secreting toxins that induce tumor cell death. Other toxins can interfere with tumor cell replication by disrupting the actin cytoskeleton and inhibiting cytokinesis. Anaerobic bacteria, particularly Clostridial spores, have the added benefit of being able to reach the oxygen-poor centers of non-resectable tumors, which are difficult for traditional chemotherapeutic agents to penetrate. There, the spores can begin growing in the hypoxic environment where they can induce tumor cell necrosis and regression of tumors.
Many of the bacteria that have shown anti-tumor efficacy are pathogenic in origin. To attenuate their pathogenicity, virulence genes can be removed or mutated, and genes involved in metabolism can also be altered to create auxotrophic organisms that can no longer replicate without human supplementation of key nutrients [7]. In the event that an attenuated bacteria does spread to the blood stream, it is critical that an effective antibiotic be used to stem the infection before serious complications such as sepsis occur.
By knowing and monitoring the susceptibility profile, clinicians will be better informed and have the confidence that—should the need arise—an effective antibiotic treatment can be administered to a patient experiencing an adverse event. To determine a bacteria’s susceptibility to a given antibiotic, a Minimum Inhibitory Concentration (MIC) assay is performed. There are numerous MIC methods, utilizing different media (e.g., broth and agar) and different systems (such as disc- or strip-based, automation, etc.). At its core, MIC assays involve testing an antimicrobial substance at different concentrations against a microbe of interest, and determining the minimum concentration of said antimicrobial that inhibits the growth of the test organisms. Once an MIC is determined, it can be compared to published clinical cut-off values for resistance or susceptibility—termed break points—for many relevant microorganism/antimicrobial combinations. In the United States, Clinical and Laboratory Standards Institute (CLSI) is the leading authority on standards for MIC test methods and break points, and is recognized by organizations such the FDA and the CDC [7,8].
While widely considered as the “gold standard” in MIC testing, the preparation of antibiotic gradients in CLSI methods means it is more time consuming to setup than other MIC methods, such as ETEST, where the antibiotic come ready-to-use in a gradient on the test strip. This makes ETEST an attractive MIC test method to developers of microbial therapeutics who wish to incorporate this method as part of their release testing and susceptibility monitoring program. A number of publications have shown that results generated using ETEST have good agreement with CLSI generated data [9,10,11]. However, differences in MIC values and major errors (e.g., MIC results indicate antibiotic resistance in one method, but is ruled susceptible in another method) have been reported as well [12,13]. A disagreement in the susceptibility of the microbial therapy to antibiotics can have dire consequences in scenarios where the therapeutic organism causes off-target infection, and an ineffective antibiotic is used due to false susceptibility test results.
Emery Pharma recently performed MIC testing for a leading microbial therapy company that wanted to determine if the ETEST method they had been using for susceptibility testing of their test organism was comparable to the reference CLSI method for a panel of 8 antibiotics. In addition to comparing the two methods, the test organism was also an auxotroph that is typically cultured in media supplemented with an amino acid mixture. To ensure that the amino acid supplement does not have antagonistic effect on the MIC test method, a 4-way comparison of ETEST vs CLSI method, and media with and without amino acid supplementation was carried out.
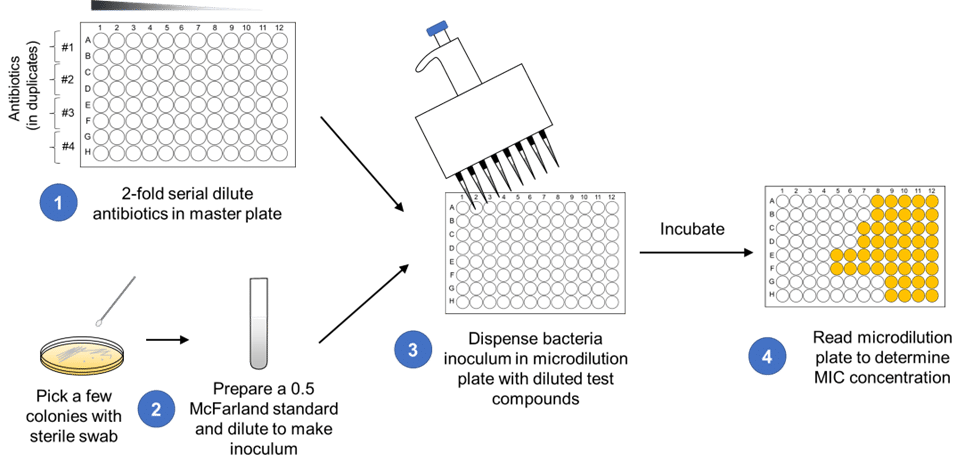
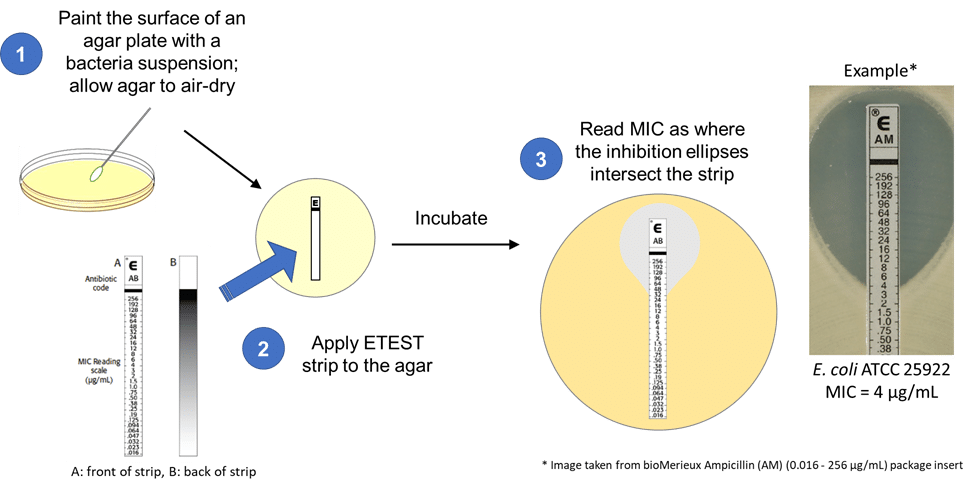
The CLSI test method used in this instance was the broth microdilution method, using 96-well microplates (schematic shown in Figure 2). The ETEST method (schematic shown in Figure 3) was run in parallel and in both methods, media with and without the amino acid supplement was assessed.
Figure 2. CLSI broth microdilution assay setup.
(Figure 3. ETEST assay setup)
When the MIC values from CLSI broth microdilution is compared to ETEST, the values for the QC strains are generally similar, differing only by around 1 Log2 value, which is accepted as the inherent variability in antimicrobial testing [15]. However, in the test strain, more than half of the antibiotics tested experienced around a 2 Log2 decrease in MIC in the ETEST method compared to the CLSI method. While this may not seem like a significant change, a higher or lower MIC can push a test strain closer or further away from the break point cutoff value for resistance. An MIC close to or past the resistance break point may affect the clinical potential of a new microbial drug with concerns for resistance holding it back from further development. It can also lead to a false sense of security, that a strain is not antibiotic resistant when tested with one methodology, when it may be far closer to the resistance break point when tested with a second method.
For these reasons, during microbial therapy development it is important to consider the AST method being used and if an alternative method is required, how its results may differ from standard methods such as those published by CLSI. If a microbial therapy is auxotrophic, the contribution of the supplement required for growth should also be assessed, in the event that the supplement interfere with the normal activity of the antibiotic.
At Emery Pharma we have a team of experienced and knowledgeable team of experts that can help guide you through the development of your microbial therapy candidate. You can reach out to us through our Contact Us page. We look forward to helping you advance the field of microbial therapeutics forward!
References:
- Sedighi M, Bialvaei AZ, Hamblin MR, Ohadi E, Asadi A, Halajzadeh M, Lohrasbi V, Mohammadzadeh N, Amiriani T, Krutova M, Amini A, Kouhsari E. Therapeutic bacteria to combat cancer; current advances, challenges, and opportunities. Cancer Med. 2019 Jun; 8(6): 3167–3181. PMID: 30950210; PMCID: PMC6558487.
- Roberts NJ, Zhang L, Janku F, Collins A, Bai RY, Staedtke V, Rusk AW, Tung D, Miller M, Roix J, Khanna KV, Murthy R, Benjamin RS, Helgason T, Szvalb AD, Bird JE, Roy-Chowdhuri S, Zhang HH, Qiao Y, Karim B, McDaniel J, Elpiner A, Sahora A, Lachowicz J, Phillips B, Turner A, Klein MK, Post G, Diaz LA Jr, Riggins GJ, Papadopoulos N, Kinzler KW, Vogelstein B, Bettegowda C, Huso DL, Varterasian M, Saha S, Zhou S. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci Transl Med. 2014 Aug 13;6(249):249ra111. doi: 10.1126/scitranslmed.3008982. PMID: 25122639; PMCID: PMC4399712.
- Vu H. Nguyen, Hyung-Seok Kim, Jung-Min Ha, Yeongjin Hong, Hyon E. Choy, Jung-Joon Min; Genetically Engineered Salmonella typhimurium as an Imageable Therapeutic Probe for Cancer. Cancer Res 1 January 2010; 70 (1): 18–23. https://doi.org/10.1158/0008-5472.CAN-09-3453
- Flickinger JC Jr, Rodeck U, Snook AE. Listeria monocytogenes as a Vector for Cancer Immunotherapy: Current Understanding and Progress. Vaccines (Basel). 2018 Jul 25;6(3):48. doi: 10.3390/vaccines6030048. PMID: 30044426; PMCID: PMC6160973.
- Kienle GS. Fever in Cancer Treatment: Coley’s Therapy and Epidemiologic Observations. Glob Adv Health Med. 2012 Mar;1(1):92-100. doi: 10.7453/gahmj.2012.1.1.016. PMID: 24278806; PMCID: PMC3833486.
- Kramer MG, Masner M, Ferreira FA, Hoffman RM. Bacterial Therapy of Cancer: Promises, Limitations, and Insights for Future Directions. Front Microbiol. 2018 Jan 23;9:16. doi: 10.3389/fmicb.2018.00016. PMID: 29472896; PMCID: PMC5810261.
- Weiman S, author; Fox J, editor. Harnessing the Power of Microbes as Therapeutics: Bugs as Drugs: Report on an American Academy of Microbiology Colloquium held in San Diego, CA, in April 2014. Washington (DC): American Society for Microbiology; 2015. Available from: https://www.ncbi.nlm.nih.gov/books/NBK519801/ doi: 10.1128/AAMCol.Apr.2014
- https://www.fda.gov/drugs/development-resources/antibacterial-susceptibility-test-interpretive-criteria
- https://www.cdc.gov/narms/antibiotics-tested.html
- Al-Hatmi AM, Normand AC, Ranque S, Piarroux R, de Hoog GS, Meletiadis J, Meis JF. Comparative Evaluation of Etest, EUCAST, and CLSI Methods for Amphotericin B, Voriconazole, and Posaconazole against Clinically Relevant Fusarium Species. Antimicrob Agents Chemother. 2016 Dec 27;61(1):e01671-16. doi: 10.1128/AAC.01671-16. PMID: 27795379; PMCID: PMC5192122.
- Ceballos-Garzon A, Garcia-Effron G, Cordoba S, Rodriguez JY, Alvarez-Moreno C, Pape PL, Parra-Giraldo CM, Morales-López S. Head-to-head comparison of CLSI, EUCAST, Etest and VITEK®2 results for Candida auris susceptibility testing. Int J Antimicrob Agents. 2022 Apr;59(4):106558. doi: 10.1016/j.ijantimicag.2022.106558. Epub 2022 Feb 25. PMID: 35227828.
- Thornsberry C, Yee YC. Comparative activity of eight antimicrobial agents against clinical bacterial isolates from the United States, measured by two methods. Am J Med. 1996 Jun 24;100(6A):26S-38S. doi: 10.1016/s0002-9343(96)00105-2. PMID: 8678094.
- van der Heijden IM, Levin AS, De Pedri EH, Fung L, Rossi F, Duboc G, Barone AA, Costa SF. Comparison of disc diffusion, Etest and broth microdilution for testing susceptibility of carbapenem-resistant P. aeruginosa to polymyxins. Ann Clin Microbiol Antimicrob. 2007 Aug 15;6:8. doi: 10.1186/1476-0711-6-8. PMID: 17697363; PMCID: PMC2018696.
- Steward CD, Mohammed JM, Swenson JM, Stocker SA, Williams PP, Gaynes RP, McGowan JE Jr, Tenover FC. Antimicrobial susceptibility testing of carbapenems: multicenter validity testing and accuracy levels of five antimicrobial test methods for detecting resistance in Enterobacteriaceae and Pseudomonas aeruginosa isolates. J Clin Microbiol. 2003 Jan;41(1):351-8. doi: 10.1128/JCM.41.1.351-358.2003. PMID: 12517872; PMCID: PMC149638.
- https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/017377s071lbl.pdf
- Jorgensen JH. Selection criteria for an antimicrobial susceptibility testing system. J Clin Microbiol. 1993 Nov;31(11):2841-4. doi: 10.1128/jcm.31.11.2841-2844.1993. PMID: 8263164; PMCID: PMC266141.