Screening for Inhibitors of Bacterial Multidrug Efflux Pumps
Microbial resistance to drug therapy is an area of increasing concern (https://www.cdc.gov/drugresistance/). The emergence of drug resistance in microorganisms can occur by several different mechanisms (Debabov, 2013), one particular mechanism being drug efflux by membrane transporter proteins, widely known as Multidrug Efflux Systems (MES). MES actively pump a wide range of drug-like molecules out of the cell, including many antimicrobial compounds.
Multidrug Efflux Systems (MES)
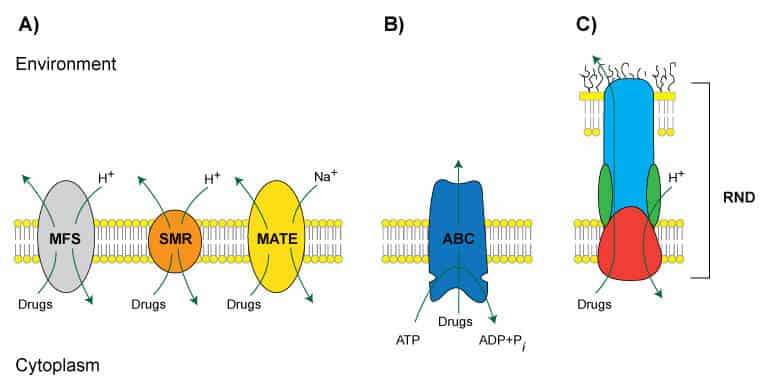
Figure 1. (Adapted from Tegos et al., 2013). Representations of known families of MES. A) The MFS, SMR, and MATE families are powered by chemiosmotic energy, pumping drugs out of the cell while pumping H+ or Na+ into the cell. B) The ABC family of pumps is powered by ATP. C) The RND family is a multisubunit complex spanning the inner and outer membranes in Gram-negative bacteria.
Efflux systems serve many essential biological roles, but they can also contribute to drug resistance. Overall, microbial MES fall into five superfamilies, namely, major facilitator (MFS), small multidrug resistance (SMR), multi-antimicrobial extrusion protein (MATE), ATP-binding cassette (ABC), and resistance-nodulation cell division (RND) (Figure 1). The natural substrates, substrate structure, and the biological functions of each superfamily vary widely and some commonly studied systems are summarized in Table 1.
|
Superfamily |
Pump(s) |
Organism |
Antibiotic substrates |
|
SMR |
Smr/QacC |
S. aureus |
QA |
|
SMR |
EmrE |
E. coli |
AC |
|
MF |
TetA |
E. coli |
TC |
|
MF |
NorA |
S. aureus |
AC, CA, FQ, PM, QA |
|
MATE |
YdhE |
E. coli |
CF, KM, NF, SM |
|
ABC |
MacB |
E. coli |
AZ, CL, EM, OL |
|
RND |
AcrB/AcrA/TolC |
E. coli |
AC, BL, CA, EM, FA, FQ, MC, NA, NV, TC |
|
RND |
MexB/MexA/OprM |
P. aeruginosa |
AZ, CF, RP |
Table 1. (Adapted from Borges-Walmsley et al., 2003). Substrate abbreviations: AC, acriflavin; AZ, azithromycin; BL, β-lactams; CA, chloramphenicol; CF, ciproflaxacin; CL, clarithromycin; EM, erythromycin; FQ, fluoroquinolones; FA, fusidic acid; KM, kanamycin; MC, mitomycin; NA, nalidixic acid; NF, norfloxacin; NV, novobiocin; OL, oleandomycin; PM, puromycin; QA, quaternary ammonium compounds (including benzalkonium chloride and cetyltrimethylammonium bromide); RP, rifampicin; SM, streptomycin; TC, tetracycline.
Studies on MES have led to the discovery of Efflux Pump Inhibitors (EPIs) that can block specific systems. Combinations of EPIs with antimicrobials may lead to the development of new therapies against drug-resistant microorganisms.
Emeryville Pharmaceutical Services offers several inhibitor assays based upon current research in MES and EPI screening techniques, and utilizes a panel of microbial strains specifically tailored to study MES. MES-knockout mutants can be used to test whether or not an applied compound is a substrate for the knocked-out MES. The application of an EPI to a MES-expressing strain should shift the Minimum Inhibitory Concentration (MIC) of a wild-type strain to the level of a MES-knockout strain.
In the example shown in Table 2, compounds 1-3 all have an MIC of 32 ug/ml against wild-type Pseudomonas aeruginosa. However, compound 1 is a strong MexB/MexA/OprM substrate with otherwise strong activity. This differs from compound 2 which is a weak substrate with intermediate activity. Compound 3 is not an efflux substrate, and it has weak activity.
|
Compound |
MIC, wild-type P. aeruginosa (ug/ml) |
MIC, ΔMexB, ΔMexA, ΔOprM P. aeruginosa (ug/ml) |
|
1 |
32 |
0.5 |
|
2 |
32 |
16 |
|
3 |
32 |
32 |
Table 2. (Posted with permission) Comparison of MICs against wild-type and efflux-knockout mutants yields information about MES specificity.
Emeryville Pharmaceutical Services also has extensive experience in developing screening methods for inhibitor interactions with specific applications. For example, MES have been implicated as a player in biofilm formation and persistence. We offer a number of techniques for growing and quantitating biofilms such as MBEC©, drip-flow reactors, and in-house biofilm reactor models, all of which can be adapted to suit any testing needs.
References
Borges-Walmsley, M. I., McKeegan, K. S., & Walmsley, A. R. (2003). Structure and function of efflux pumps that confer resistance to drugs. The Biochemical Journal, 376, 313–338.
Debabov, D. (2013). Antibiotic Resistance: Origins, Mechanisms, Approaches to Counter. Applied Biochemistry and Microbiology, 49, 1–7.
Tegos, G. P., Haynes, M., Strouse, J. J., Khan, M. T., Bologa, C. G., Oprea, T. I., & Sklar, L. A. (2013). Curr Pharm Des, 17, 1291–1302.
